Update Data
Time to update your data file with new data. Download this data and append it to data/data.csv in your repo, being sure not to duplicate the header row:
| period | BA | DM | DO | DS | NA | OL | OT | PB | PE | PF | PH | PI | PL | PM | PP | RF | RM | RO | SF | SH | SO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 451 | 0 | 35 | 12 | 1 | 1 | 2 | 2 | 4 | 0 | 5 | 0 | 0 | 0 | 0 | 84 | 0 | 0 | 0 | 1 | 0 | 0 |
| 452 | 0 | 30 | 12 | 1 | 1 | 1 | 4 | 6 | 3 | 2 | 0 | 0 | 0 | 0 | 93 | 0 | 0 | 0 | 0 | 0 | 0 |
| 453 | 0 | 34 | 9 | 0 | 3 | 2 | 4 | 6 | 5 | 1 | 0 | 0 | 0 | 0 | 76 | 0 | 0 | 0 | 0 | 0 | 0 |
| 454 | 0 | 34 | 5 | 1 | 3 | 1 | 7 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 38 | 0 | 0 | 0 | 0 | 0 | 0 |
| 455 | 0 | 42 | 6 | 1 | 4 | 2 | 10 | 5 | 5 | 0 | 0 | 0 | 1 | 0 | 9 | 0 | 0 | 0 | 0 | 1 | 0 |
| 458 | 0 | 61 | 16 | 1 | 5 | 1 | 9 | 1 | 4 | 0 | 0 | 0 | 4 | 3 | 0 | 0 | 11 | 2 | 0 | 3 | 0 |
| 459 | 0 | 62 | 20 | 1 | 7 | 0 | 6 | 2 | 6 | 0 | 0 | 0 | 5 | 2 | 2 | 1 | 10 | 2 | 0 | 1 | 0 |
| 460 | 0 | 55 | 17 | 0 | 0 | 3 | 11 | 1 | 11 | 0 | 0 | 0 | 3 | 4 | 44 | 0 | 10 | 4 | 0 | 0 | 0 |
| 461 | 0 | 63 | 19 | 0 | 6 | 2 | 8 | 2 | 11 | 1 | 0 | 0 | 2 | 1 | 44 | 1 | 1 | 0 | 0 | 8 | 0 |
| 4620 | 0 | 44 | 24 | 0 | 5 | 1 | 11 | 5 | 10 | 1 | 1 | 0 | 1 | 0 | 92 | 0 | 1 | 1 | 0 | 0 | 0 |
| 463 | 0 | 33 | 7 | 1 | 7 | 0 | 8 | 2 | 2 | 1 | 2 | 0 | 0 | 0 | 108 | 0 | 0 | 0 | 0 | 2 | 0 |
| 465 | 0 | 33 | 9 | 0 | 1 | 0 | 15 | 8 | 2 | 0 | 1 | 0 | 0 | 0 | 158 | 1 | 0 | 0 | 0 | 0 | 0 |
| 466 | 0 | 42 | 7 | 0 | 6 | 0 | 15 | 6 | 1 | 0 | 1 | 0 | 0 | 0 | 213 | 0 | 0 | 0 | 0 | 0 | 0 |
| 467 | 0 | 41 | 5 | 1 | 6 | 0 | 26 | 5 | 9 | 0 | 2 | 0 | 1 | 1 | 94 | 0 | 1 | 0 | 0 | 1 | 0 |
This is more rodent abundance data from Portal, from more recent sampling periods.
-
Add, commit, and push your changes.
-
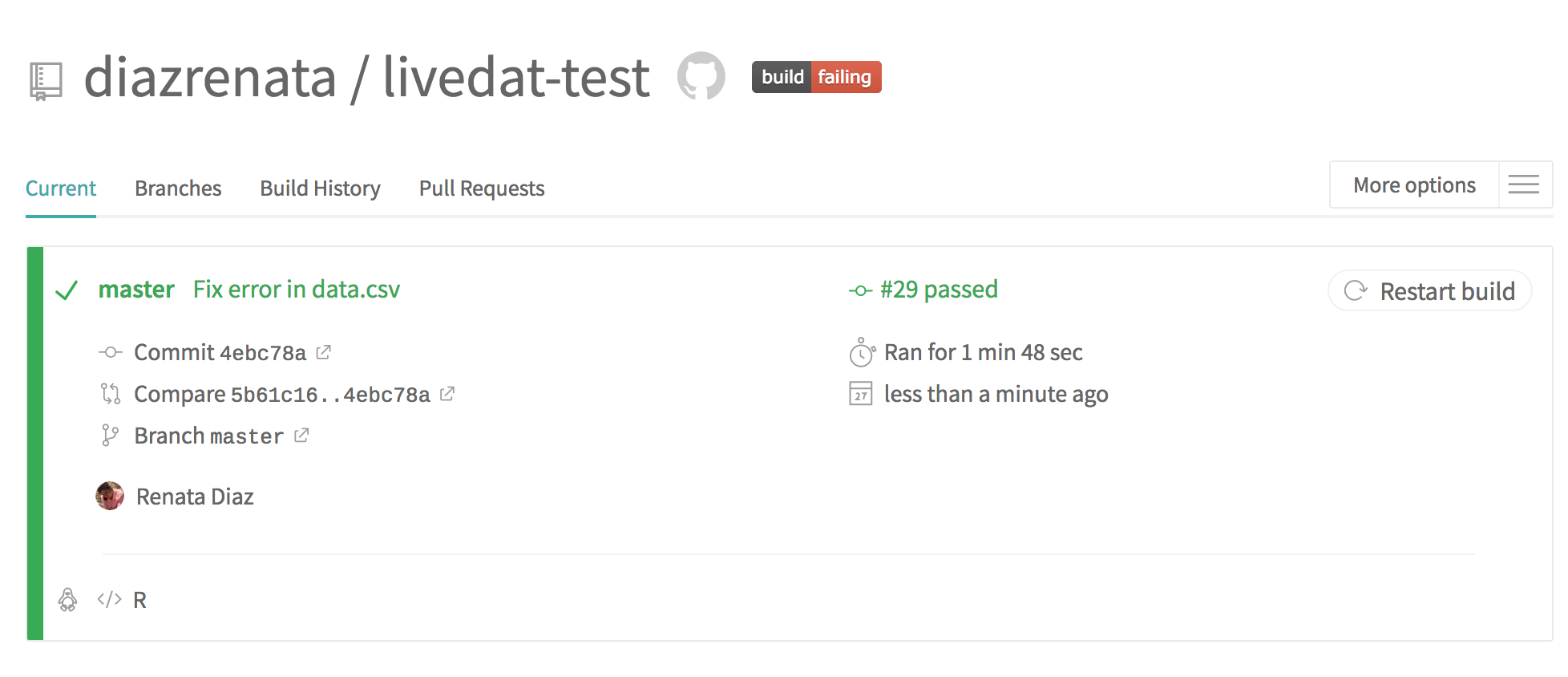
Check that your changes are on GitHub and that your tests are passing on Travis. (It may take a few moments for your checks to run).
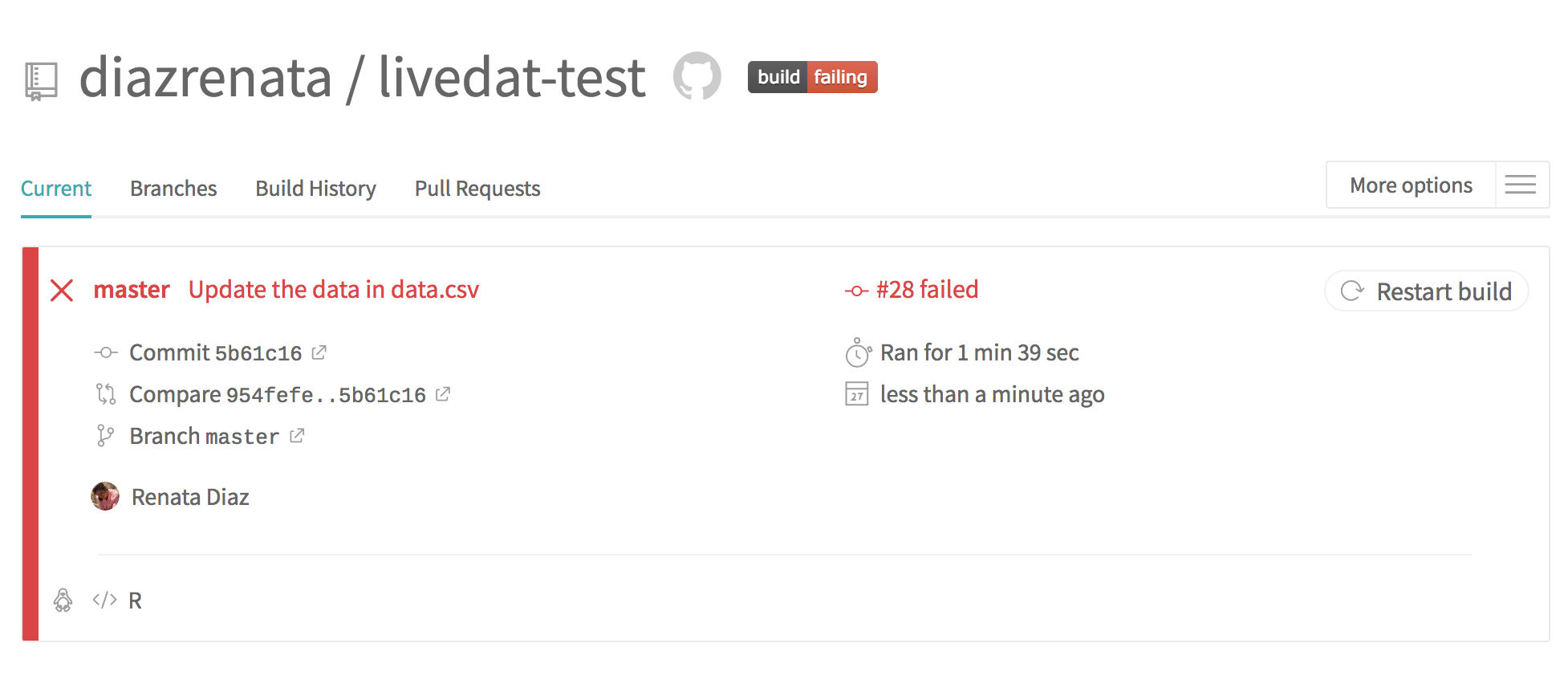 The data we just added caused the tests to fail! Specifically, the period values contain an error.
The data we just added caused the tests to fail! Specifically, the period values contain an error.
-
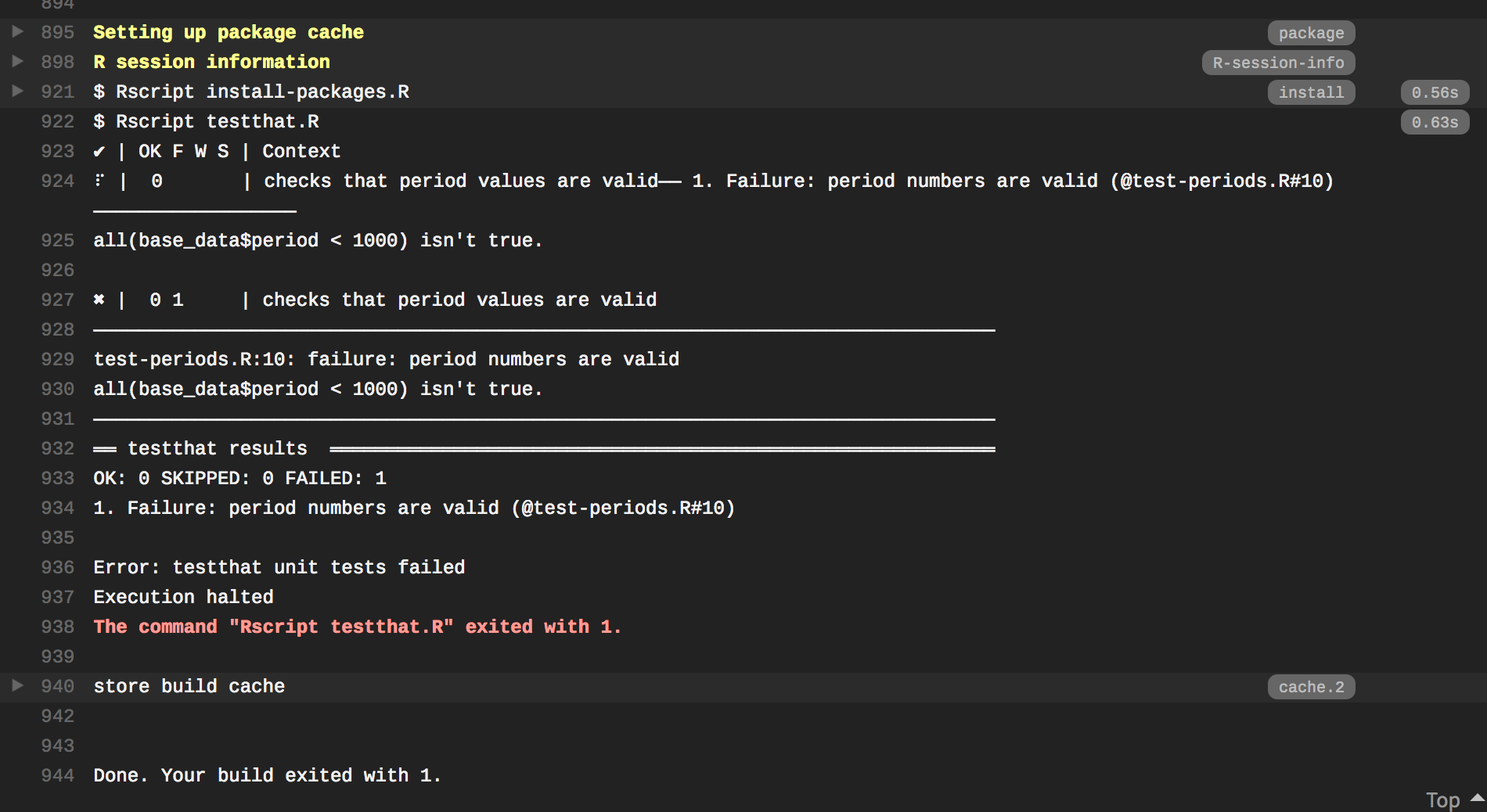
Let’s open our data in R to find the lines with the error.
rodent_data <- read.csv('data/data.csv')
rodent_data[ which(rodent_data$period > 1000), ]
> period BA DM DO DS NA. OL OT PB PE PF PH PI PL PM PP RF RM RO SF SH SO
20 4620 0 44 24 0 5 1 11 5 10 1 1 0 1 0 92 0 1 1 0 0 0
- On line 20, the period is
4620. Correct this to462and re-save the data:
rodent_data$period[20] <- 462
write.csv(rodent_data, 'data/data.csv', row.names = F)
-
Add, commit, and push your results.
-
Go to GitHub and Travis and check that the new data has been added and the tests are all now passing.